Yue WU
Doctorante Université
novembre 2019 - octobre 2023
| Équipe : |
Thèmes de recherche
Characterization of diatom Phaeodactylum tricornutum epigenetic code & its role in response to the environment: extending the histone code and analyzing its crosstalk
Projets
Parcours universitaire
* 2014 – 2017 : Master in Environmental Engineering (University of Chinese Academy of Sciences, Institute of Hydrobiology)
* 2019- present: PhD in Epigenomics , University of Nantes
Publications
1 publication
Timothée, Chaumier; Yang, Feng; Manirakiza, Eric; Ait-Mohamed, Ouardia; Wu, Yue; Chandola, Udita; Jesus, Bruno; Piganeau, Gwenael; Groisillier, Agnès; Tirichine, Leila
Genome-wide assessment of genetic diversity and transcript variations in 17 accessions of the model diatom Phaeodactylum tricornutum Article de journal
Dans: ISME Communications, vol. 4, iss. 1, p. ycad008, 2024.
@article{Timothée2024,
title = {Genome-wide assessment of genetic diversity and transcript variations in 17 accessions of the model diatom Phaeodactylum tricornutum},
author = {Chaumier Timothée and Feng Yang and Eric Manirakiza and Ouardia Ait-Mohamed and Yue Wu and Udita Chandola and Bruno Jesus and Gwenael Piganeau and Agnès Groisillier and Leila Tirichine},
url = {https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10833087/},
doi = {10.1093/ismeco/ycad008},
year = {2024},
date = {2024-01-10},
urldate = {2024-01-10},
journal = {ISME Communications},
volume = {4},
issue = {1},
pages = {ycad008},
abstract = {Diatoms, a prominent group of phytoplankton, have a significant impact on both the oceanic food chain and carbon sequestration, thereby playing a crucial role in regulating the climate. These highly diverse organisms show a wide geographic distribution across various latitudes. In addition to their ecological significance, diatoms represent a vital source of bioactive compounds that are widely used in biotechnology applications. In the present study, we investigated the genetic and transcriptomic diversity of 17 accessions of the model diatom Phaeodactylum tricornutum including those sampled a century ago as well as more recently collected accessions. The analysis of the data reveals a higher genetic diversity and the emergence of novel clades, indicating an increasing diversity within the P. tricornutum population structure, compared to the previous study and a persistent long-term balancing selection of genes in old and newly sampled accessions. However, the study did not establish a clear link between the year of sampling and genetic diversity, thereby, rejecting the hypothesis of loss of heterozygoty in cultured strains. Transcript analysis identified novel transcript including noncoding RNA and other categories of small RNA such as PiwiRNAs. Additionally, transcripts analysis using differential expression as well as Weighted Gene Correlation Network Analysis has provided evidence that the suppression or downregulation of genes cannot be solely attributed to loss-of-function mutations. This implies that other contributing factors, such as epigenetic modifications, may play a crucial role in regulating gene expression. Our study provides novel genetic resources, which are now accessible through the platform PhaeoEpiview (https://PhaeoEpiView.univ-nantes.fr), that offer both ease of use and advanced tools to further investigate microalgae biology and ecology, consequently enriching our current understanding of these organisms.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
3 publications
Wu, Yue; Tirichine, Leila
Chromosome-Wide Distribution and Characterization of H3K36me3 and H3K27Ac in the Marine Model Diatom Phaeodactylum tricornutum Article de journal
Dans: Plants (Basel), vol. 12, iss. 15, p. 2852, 2023.
@article{Wu2023b,
title = {Chromosome-Wide Distribution and Characterization of H3K36me3 and H3K27Ac in the Marine Model Diatom Phaeodactylum tricornutum},
author = {Yue Wu and Leila Tirichine },
url = {https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10421102/
hal-04284555v1 },
doi = { 10.3390/plants12152852},
year = {2023},
date = {2023-08-02},
urldate = {2023-08-02},
journal = {Plants (Basel)},
volume = {12},
issue = {15},
pages = {2852},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Hisanaga, Tetsuya; Romani, Facundo; Wu, Shuangyang; Kowar, Teresa; Wu, Yue; Lintermann, Ruth; Fridrich, Arie; Cho, Chung Hyun; Chaumier, Timothée; Jamge, Bhagyshree; Montgomery, Sean A; Axelsson, Elin; Akimcheva, Svetlana; Dierschke, Tom; Bowman, John L; Fujiwara, Takayuki; Hirooka, Shunsuke; Miyagishima, Shin-Ya; Dolan, Liam; Tirichine, Leila; Schubert, Daniel; Berger, Frédéric
The Polycomb repressive complex 2 deposits H3K27me3 and represses transposable elements in a broad range of eukaryotes Article de journal
Dans: Current Biology, vol. 33, iss. 20, p. 4367-4380.e9, 2023.
@article{Hisanaga2023,
title = {The Polycomb repressive complex 2 deposits H3K27me3 and represses transposable elements in a broad range of eukaryotes},
author = {Tetsuya Hisanaga and Facundo Romani and Shuangyang Wu and Teresa Kowar and Yue Wu and Ruth Lintermann and Arie Fridrich and Chung Hyun Cho and Timothée Chaumier and Bhagyshree Jamge and Sean A Montgomery and Elin Axelsson and Svetlana Akimcheva and Tom Dierschke and John L Bowman and Takayuki Fujiwara and Shunsuke Hirooka and Shin-Ya Miyagishima and Liam Dolan and Leila Tirichine and Daniel Schubert and Frédéric Berger},
url = {https://www.sciencedirect.com/science/article/pii/S0960982223011533?via%3Dihub
hal-04284522v1},
doi = {10.1016/j.cub.2023.08.073},
year = {2023},
date = {2023-08-02},
urldate = {2023-08-02},
journal = {Current Biology},
volume = {33},
issue = {20},
pages = {4367-4380.e9},
abstract = {The mobility of transposable elements (TEs) contributes to evolution of genomes. Their uncontrolled activity causes genomic instability; therefore, expression of TEs is silenced by host genomes. TEs are marked with DNA and H3K9 methylation, which are associated with silencing in flowering plants, animals, and fungi. However, in distantly related groups of eukaryotes, TEs are marked by H3K27me3 deposited by the Polycomb repressive complex 2 (PRC2), an epigenetic mark associated with gene silencing in flowering plants and animals. The direct silencing of TEs by PRC2 has so far only been shown in one species of ciliates. To test if PRC2 silences TEs in a broader range of eukaryotes, we generated mutants with reduced PRC2 activity and analyzed the role of PRC2 in extant species along the lineage of Archaeplastida and in the diatom P. tricornutum. In this diatom and the red alga C. merolae, a greater proportion of TEs than genes were repressed by PRC2, whereas a greater proportion of genes than TEs were repressed by PRC2 in bryophytes. In flowering plants, TEs contained potential cis-elements recognized by transcription factors and associated with neighbor genes as transcriptional units repressed by PRC2. Thus, silencing of TEs by PRC2 is observed not only in Archaeplastida but also in diatoms and ciliates, suggesting that PRC2 deposited H3K27me3 to silence TEs in the last common ancestor of eukaryotes. We hypothesize that during the evolution of Archaeplastida, TE fragments marked with H3K27me3 were selected to shape transcriptional regulation, controlling networks of genes regulated by PRC2.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Wu, Yue; Chaumier, Timothée; Manirakiza, Eric; Veluchamy, Alaguraj; Tirichine, Leila
PhaeoEpiView: an epigenome browser of the newly assembled genome of the model diatom Phaeodactylum tricornutum Article de journal
Dans: Scientific Reports , vol. 13, p. 8320, 2023, ISSN: 2045-2322.
@article{Wu2023,
title = {PhaeoEpiView: an epigenome browser of the newly assembled genome of the model diatom Phaeodactylum tricornutum},
author = {Yue Wu and Timothée Chaumier and Eric Manirakiza and Alaguraj Veluchamy and Leila Tirichine},
url = {https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10206091/
hal-04284562v1 },
doi = { 10.1038/s41598-023-35403-1},
issn = {2045-2322},
year = {2023},
date = {2023-05-23},
urldate = {2023-05-23},
journal = {Scientific Reports },
volume = {13},
pages = {8320},
abstract = {Recent advances in DNA sequencing technologies particularly long-read sequencing, greatly improved genomes assembly. However, this has created discrepancies between published annotations and epigenome tracks, which have not been updated to keep pace with the new assemblies. Here, we used the latest improved telomere-to-telomere assembly of the model pennate diatom Phaeodactylum tricornutum to lift over the gene models from Phatr3, a previously annotated reference genome. We used the lifted genes annotation and newly published transposable elements to map the epigenome landscape, namely DNA methylation and post-translational modifications of histones. This provides the community with PhaeoEpiView, a browser that allows the visualization of epigenome data and transcripts on an updated and contiguous reference genome, to better understand the biological significance of the mapped data. We updated previously published histone marks with a more accurate peak calling using mono instead of poly(clonal) antibodies and deeper sequencing. PhaeoEpiView (https://PhaeoEpiView.univ-nantes.fr) will be continuously updated with the newly published epigenomic data, making it the largest and richest epigenome browser of any stramenopile. In the upcoming era of molecular environmental studies, where epigenetics plays a significant role, we anticipate that PhaeoEpiView will become a widely used tool.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1 publication
Wu, Yue; Timothée, Chaumier; Manirakiza, Eric; Veluchamy, Alaguraj; Tirichine, Leila
PhaeoEpiView: An epigenome browser of the newly assembled genome of the model diatom Phaeodactylum tricornutum Article de journal À paraître
Dans: bioRxiv, À paraître.
@article{Wu2022.07.29.502047,
title = {PhaeoEpiView: An epigenome browser of the newly assembled genome of the model diatom Phaeodactylum tricornutum},
author = {Yue Wu and Chaumier Timothée and Eric Manirakiza and Alaguraj Veluchamy and Leila Tirichine},
url = {https://www.biorxiv.org/content/early/2022/08/01/2022.07.29.502047},
doi = {10.1101/2022.07.29.502047},
year = {2022},
date = {2022-08-01},
urldate = {2022-01-01},
journal = {bioRxiv},
publisher = {Cold Spring Harbor Laboratory},
abstract = {Motivation Recent advances in DNA sequencing technologies in particular of long reads type greatly improved genomes assembly leading to discrepancies between both published annotations and epigenome tracks which did not keep pace with new assemblies. This comprises the availability of accurate resources which penalizes the progress in research.Results Here, we used the latest improved telomere to telomere assembly of the model pennate diatom Phaeodactylum tricornutum to lift over the gene models from Phatr3, a previously annotated reference genome. We used the lifted genome annotation including genes and transposable elements to map the epigenome landscape, namely DNA methylation and post translational modifications of histones providing the community with PhaeoEpiView, a browser that allows the visualization of epigenome data as well as transcripts on an updated reference genome to better understand the biological significance of the mapped data on contiguous genome rather than a fragmented one. We updated previously published histone marks with a more accurate mapping using monoclonal antibodies instead of polyclonal and deeper sequencing. PhaeoEpiView will be continuously updated with the newly published epigenomic data making it the largest and richest epigenome browser of any stramenopile. We expect that PhaeoEpiView will be a standard tool for the coming era of molecular environmental studies where epigenetics holds a place of choice.Availability PhaeoEpiView is available at: https://PhaeoEpiView.univ-nantes.frCompeting Interest StatementThe authors have declared no competing interest.},
keywords = {},
pubstate = {forthcoming},
tppubtype = {article}
}
1 publication
Bourdareau, Simon; Tirichine, Leila; Lombard, Bérangère; Loew, Damarys; Scornet, Delphine; Wu, Yue; Coelho, Susana M; Cock, Mark J
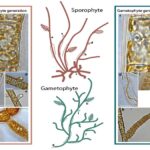
Histone modifications during the life cycle of the brown alga Ectocarpus Article de journal
Dans: Genome Biology, vol. 22, no. 1, 2021, ISSN: 1474760X.
@article{Bourdareau2021,
title = {Histone modifications during the life cycle of the brown alga Ectocarpus},
author = {Simon Bourdareau and Leila Tirichine and Bérangère Lombard and Damarys Loew and Delphine Scornet and Yue Wu and Susana M Coelho and Mark J Cock},
url = {https://pubmed.ncbi.nlm.nih.gov/33397407/},
doi = {10.1186/s13059-020-02216-8},
issn = {1474760X},
year = {2021},
date = {2021-12-01},
journal = {Genome Biology},
volume = {22},
number = {1},
publisher = {BioMed Central Ltd},
abstract = {Background: Brown algae evolved complex multicellularity independently of the animal and land plant lineages and are the third most developmentally complex phylogenetic group on the planet. An understanding of developmental processes in this group is expected to provide important insights into the evolutionary events necessary for the emergence of complex multicellularity. Here, we focus on mechanisms of epigenetic regulation involving post-translational modifications of histone proteins. Results: A total of 47 histone post-translational modifications are identified, including a novel mark H2AZR38me1, but Ectocarpus lacks both H3K27me3 and the major polycomb complexes. ChIP-seq identifies modifications associated with transcription start sites and gene bodies of active genes and with transposons. H3K79me2 exhibits an unusual pattern, often marking large genomic regions spanning several genes. Transcription start sites of closely spaced, divergently transcribed gene pairs share a common nucleosome-depleted region and exhibit shared histone modification peaks. Overall, patterns of histone modifications are stable through the life cycle. Analysis of histone modifications at generation-biased genes identifies a correlation between the presence of specific chromatin marks and the level of gene expression. Conclusions: The overview of histone post-translational modifications in the brown alga presented here will provide a foundation for future studies aimed at understanding the role of chromatin modifications in the regulation of brown algal genomes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}