Stagiaire M2 sur les interactions plantes - plantes - microorganismes
| Durée du contrat : | 6 mois |
| Début du contrat : | lundi 05 janvier 2026 |
| Type de contrat : | stage |
| Rémunération : | 650 € brut mensuel |
| Équipe de rattachement : | Interactions plante-plante et signaux rhizosphériques |
Missions :
– Caractérisation d’une communauté synthétique microbienne (SynCom) par des approches de microbiologie, test in vitro sur les étapes précoces du cycle de P. ramosa.
– Test d’inoculation de la SynCom dans le système ExuFlow (culture miniaturisée de 24 plantes d’A. thaliana avec recirculation du milieu d’arrosage), suivi d’analyses sur trois semaines. Les plantules sauvages Col-0 seront transférées et inoculées en semaine 0. Chaque semaine, un phénotypage sera réalisé et les exsudats collectés pour des analyses métabolomiques bioguidées. En fin de culture, les plantes et leur rhizosphère seront récoltées pour des analyses futures (transcriptomique et métabarcoding). L’expérience pourra être conduite plusieurs fois pour avoir des répliquas de culture.
Environnement et contexte de travail :
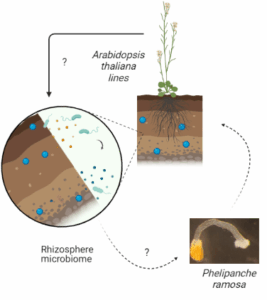
Les plantes libèrent une variété de métabolites dans la rhizosphère, influençant les organismes environnants, y compris d’autres plantes et les micro-organismes. Parmi ces métabolites, les strigolactones et les glucosinolates (ainsi que leurs dérivés, comme les isothiocyanates) jouent un rôle important. Ces métabolites secondaires peuvent induire la germination de plantes parasites comme l’orobanche rameuse (P. ramosa) et modifier la signalisation moléculaire avec les micro-organismes. En retour, le microbiome de la rhizosphère peut influencer le succès des plantes parasites.
Dans le cadre de ce projet, l’objectif est d’étudier l’effet d’une communauté synthétique microbienne (SynCom) à activité ‘stimulatrice’ sur le phénotype et la physiologie ainsi que sur l’exsudome de la plante hôte A. thaliana Col-0, et donc sur l’activité de germination de la plante parasite associée P. ramosa.
Références :
Auger, B. Mol. Plant-Microbe Interact. 25, 993–1004 (2012). Accès libre*.
Martinez, L. Plant and Soil 483 (1), 667–691 (2023). Accès libre*.
Activités principales :
– phénotypage : mesures de l’appareil végétatif (diamètre de la rosette, nombre de feuilles, taille de la plante) et photographies standardisées de l’état phénotypique, et stade phénologique (stade végétatif rosette ou tige, floraison, fructification)
– récolte/analyse d’exsudats : tests de germination de P. ramosa, et analyse bioguidée pour caractériser les métabolites d’intérêt par métabolomique ciblée.
– récolte racines et rhizosphere, extraction ARN/ADN pour des analyses transcriptomiques et métabarcoding rhizosphère. Si le temps le permet les données issues du séquençage de ces analyses pourront être étudiées.
Compétences et profil recherché :
– Connaissances souhaitées en écologie microbienne ou biologie végétale ou biochimie.
– Expérience en laboratoire biologie végétale, microbiologie, biologie moléculaire…
– Bonne maîtrise de l’analyse de données, des biostatistiques (R/rstudio)
– Fort intérêt pour la recherche académique/fondamentale.
Autres informations générales :
Veuillez envoyer votre CV et lettre de motivation à lucie.poulin@univ-nantes.fr et ahmed.choukri@univ-nantes.fr
Date limite de réception des candidatures : mercredi 01 octobre 2025
Contact : Lucie POULIN , Ahmed CHOUKRI ,