Responsable : Cathy CHARLIER

Les activités de la plateforme Interactions Moléculaires Puces Activités (IMPACT) se positionnent en protéomique fonctionnelle.
La plateforme IMPACT propose un ensemble d’approches pour comprendre la façon dont les molécules biologiques et chimiques interagissent entre elles afin d’en décrypter leur fonction.
Savoir-faire
La plateforme IMPACT offre une gamme de technologies dans 4 domaines d’expertise :
- L’analyse de profils d’abondance protéique ou de modifications post-traductionnelles avec des puces à protéines dédiées (protein microarray, SPRimaging) et des technologies multiparamétriques (label free, alphaLISA).
- Le criblage de molécules (protéines, peptides, molécules chimiques…) capables de moduler des interactions (iTC, AlphaLISA).
- La caractérisation des paramètres d’interaction moléculaire en solution ou aux interfaces grâce à des technologies analytiques (SPR, BLItz, iTC200).
- Le traitement et l’intégration des données issues des profils d’expression ou du criblage grâce à des analyses bio-informatiques (Galaxy).

La plateforme répond aux besoins des laboratoires académiques et privés dans les domaines de la santé, de l’agro-alimentaire, de la mer et des biotechnologies.
![]()
Mots-clés : AlphaLISA, dichroïsme circulaire, interférométrie, microcalorimétrie, puces à protéines, spectrofluorimétrie
Membres
Publications
1 publication
Nifontova, Galina; Charlier, Cathy; Ayadi, Nizar; Fleury, Fabrice; Karaulov, Alexander; Sukhanova, Alyona; Nabiev, Igor
Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate Article de journal
Dans: Biosensors, vol. 14, no. 1, 2024, ISSN: 2079-6374.
@article{bios14010043,
title = {Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate},
author = {Galina Nifontova and Cathy Charlier and Nizar Ayadi and Fabrice Fleury and Alexander Karaulov and Alyona Sukhanova and Igor Nabiev},
url = {https://www.mdpi.com/2079-6374/14/1/43
hal-04449485v1 },
doi = {10.3390/bios14010043},
issn = {2079-6374},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Biosensors},
volume = {14},
number = {1},
abstract = {Photonic crystals (PCs) are promising tools for label-free sensing in drug discovery screening, diagnostics, and analysis of ligand-receptor interactions. Imaging of PC surface modes has emerged as a novel approach to the detection of multiple binding events at the sensor surface. PC surface modification and decoration with recognition units yield an interface providing the highly sensitive detection of cancer biomarkers, antibodies, and oligonucleotides. The RAD51 protein plays a central role in DNA repair via the homologous recombination pathway. This recombinase is essential for the genome stability and its overexpression is often correlated with aggressive cancer. RAD51 is therefore a potential target in the therapeutic strategy for cancer. Here, we report the designing of a PC-based array sensor for real-time monitoring of oligonucleotide-RAD51 recruitment by means of surface mode imaging and validation of the concept of this approach. Our data demonstrate that the designed biosensor ensures the highly sensitive multiplexed analysis of association-dissociation events and detection of the biomarker of DNA damage using a microfluidic PC array. The obtained results highlight the potential of the developed technique for testing the functionality of candidate drugs, discovering new molecular targets and drug entities. This paves the way to further adaption and bioanalytical use of the biosensor for high-content screening to identify new DNA repair inhibitor drugs targeting the RAD51 nucleoprotein filament or to discover new molecular targets.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2 publications
Mauro, E; Lapaillerie, D; Tumiotto, C; Charlier, Cathy; Martins, F; Sousa, S F; Métifiot, M; Weigel, Pierre; Yamatsugu, K; Kanai, M; Munier-Lehmann, H; Richetta, C; Maisch, M; Dutrieux, J; Batisse, J; Ruff, M; Delelis, O; Lesbats, P; Parissi, V
Modulation of the functional interfaces between retroviral intasomes and the human nucleosome Article de journal
Dans: mBio, p. e0108323, 2023, ISSN: 2150-7511.
@article{pmid37382440,
title = {Modulation of the functional interfaces between retroviral intasomes and the human nucleosome},
author = {E Mauro and D Lapaillerie and C Tumiotto and Cathy Charlier and F Martins and S F Sousa and M Métifiot and Pierre Weigel and K Yamatsugu and M Kanai and H Munier-Lehmann and C Richetta and M Maisch and J Dutrieux and J Batisse and M Ruff and O Delelis and P Lesbats and V Parissi},
doi = {10.1128/mbio.01083-23},
issn = {2150-7511},
year = {2023},
date = {2023-06-01},
urldate = {2023-06-01},
journal = {mBio},
pages = {e0108323},
abstract = {Infection by retroviruses as HIV-1 requires the stable integration of their genome into the host cells. This process needs the formation of integrase (IN)-viral DNA complexes, called intasomes, and their interaction with the target DNA wrapped around nucleosomes within cell chromatin. To provide new tools to analyze this association and select drugs, we applied the AlphaLISA technology to the complex formed between the prototype foamy virus (PFV) intasome and nucleosome reconstituted on 601 Widom sequence. This system allowed us to monitor the association between both partners and select small molecules that could modulate the intasome/nucleosome association. Using this approach, drugs acting either on the DNA topology within the nucleosome or on the IN/histone tail interactions have been selected. Within these compounds, doxorubicin and histone binders calixarenes were characterized using biochemical, molecular simulations and cellular approaches. These drugs were shown to inhibit both PFV and HIV-1 integration . Treatment of HIV-1-infected PBMCs with the selected molecules induces a decrease in viral infectivity and blocks the integration process. Thus, in addition to providing new information about intasome-nucleosome interaction determinants, our work also paves the way for further unedited antiviral strategies that target the final step of intasome/chromatin anchoring. IMPORTANCE In this work, we report the first monitoring of retroviral intasome/nucleosome interaction by AlphaLISA. This is the first description of the AlphaLISA application for large nucleoprotein complexes (>200 kDa) proving that this technology is suitable for molecular characterization and bimolecular inhibitor screening assays using such large complexes. Using this system, we have identified new drugs disrupting or preventing the intasome/nucleosome complex and inhibiting HIV-1 integration both and in infected cells. This first monitoring of the retroviral/intasome complex should allow the development of multiple applications including the analyses of the influence of cellular partners, the study of additional retroviral intasomes, and the determination of specific interfaces. Our work also provides the technical bases for the screening of larger libraries of drugs targeting specifically these functional nucleoprotein complexes, or additional nucleosome-partner complexes, as well as for their characterization.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Nifontova, Galina; Petrova, Irina; Gerasimovich, Evgeniia; Konopsky, Valery N.; Ayadi, Nizar; Charlier, Cathy; Fleury, Fabrice; Karaulov, Alexander; Sukhanova, Alyona; Nabiev, Igor
Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging Article de journal
Dans: International Journal of Molecular Sciences, vol. 24, no. 5, 2023, ISSN: 1422-0067.
@article{ijms24054347b,
title = {Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging},
author = {Galina Nifontova and Irina Petrova and Evgeniia Gerasimovich and Valery N. Konopsky and Nizar Ayadi and Cathy Charlier and Fabrice Fleury and Alexander Karaulov and Alyona Sukhanova and Igor Nabiev},
url = {https://www.mdpi.com/1422-0067/24/5/4347},
doi = {10.3390/ijms24054347},
issn = {1422-0067},
year = {2023},
date = {2023-02-22},
urldate = {2023-02-22},
journal = {International Journal of Molecular Sciences},
volume = {24},
number = {5},
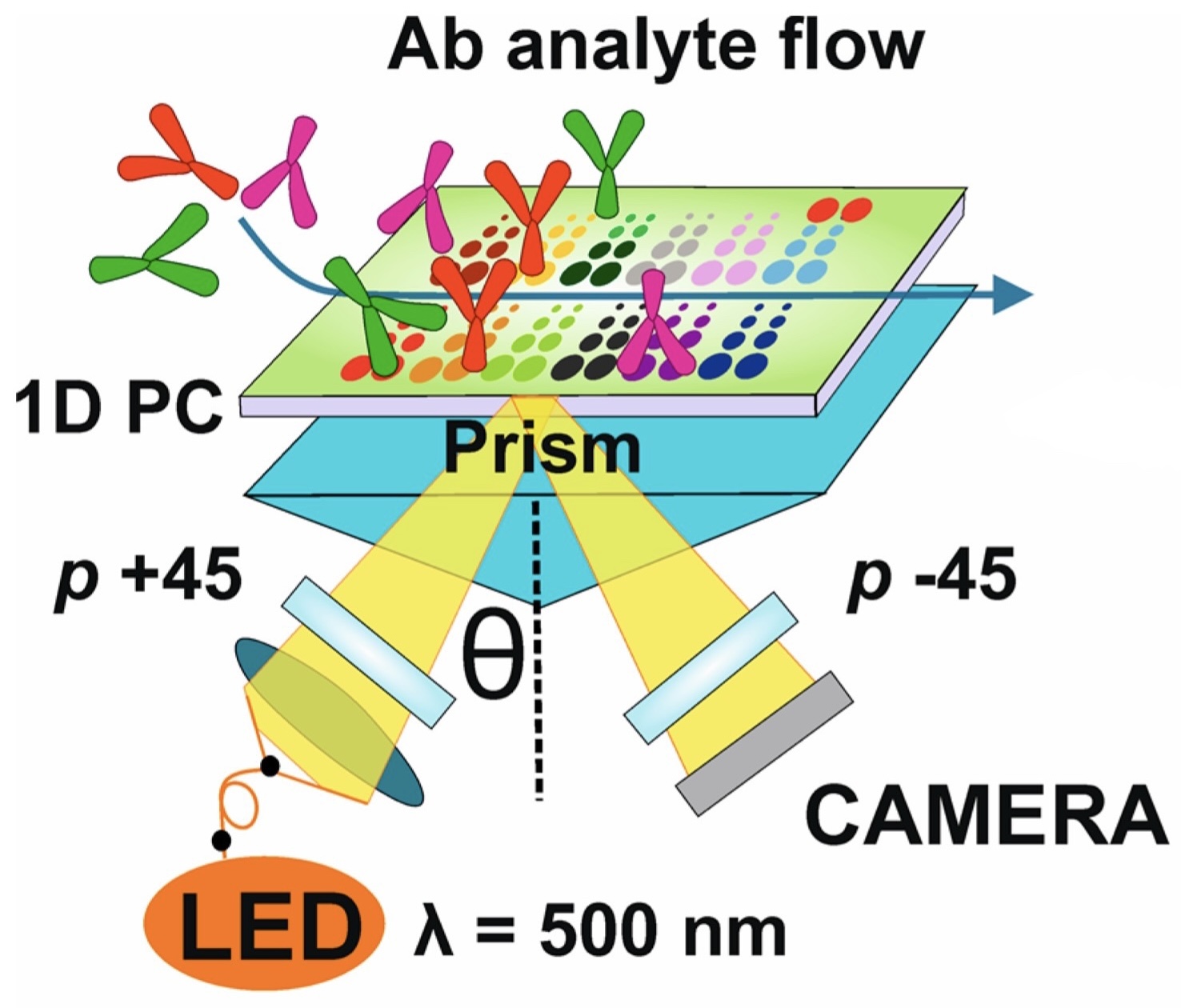
abstract = {High-throughput protein assays are crucial for modern diagnostics, drug discovery, proteomics, and other fields of biology and medicine. It allows simultaneous detection of hundreds of analytes and miniaturization of both fabrication and analytical procedures. Photonic crystal surface mode (PC SM) imaging is an effective alternative to surface plasmon resonance (SPR) imaging used in conventional gold-coated, label-free biosensors. PC SM imaging is advantageous as a quick, label-free, and reproducible technique for multiplexed analysis of biomolecular interactions. PC SM sensors are characterized by a longer signal propagation at the cost of a lower spatial resolution, which makes them more sensitive than classical SPR imaging sensors. We describe an approach for designing label-free protein biosensing assays employing PC SM imaging in the microfluidic mode. Label-free, real-time detection of PC SM imaging biosensors using two-dimensional imaging of binding events has been designed to study arrays of model proteins (antibodies, immunoglobulin G-binding proteins, serum proteins, and DNA repair proteins) at 96 points prepared by automated spotting. The data prove feasibility of simultaneous PC SM imaging of multiple protein interactions. The results pave the way to further develop PC SM imaging as an advanced label-free microfluidic assay for the multiplexed detection of protein interactions.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1 publication
Lapaillerie, Delphine; Charlier, Cathy; Guyonnet-Dupérat, Véronique; Murigneux, Emilie; Fernandes, Henrique S.; Martins, Fábio G.; Magalhães, Rita P.; Vieira, Tatiana F.; Richetta, Clémence; Subra, Frédéric; Lebourgeois, Samuel; Charpentier, Charlotte; Descamps, Diane; Visseaux, Benoît; Weigel, Pierre; Favereaux, Alexandre; Beauvineau, Claire; Buron, Frédéric; Teulade-Fichou, Marie-Paule; Routier, Sylvain; Gallois-Montbrun, Sarah; Meertens, Laurent; Delelis, Olivier; Sousa, Sérgio F.; Parissi, Vincent
Selection of Bis-Indolyl Pyridines and Triphenylamines as New Inhibitors of SARS-CoV-2 Cellular Entry by Modulating the Spike Protein/ACE2 Interfaces Article de journal
Dans: Antimicrobial Agents and Chemotherapy, vol. 0, no. 0, p. e00083-22, 2022.
@article{doi:10.1128/aac.00083-22,
title = {Selection of Bis-Indolyl Pyridines and Triphenylamines as New Inhibitors of SARS-CoV-2 Cellular Entry by Modulating the Spike Protein/ACE2 Interfaces},
author = {Delphine Lapaillerie and Cathy Charlier and Véronique Guyonnet-Dupérat and Emilie Murigneux and Henrique S. Fernandes and Fábio G. Martins and Rita P. Magalhães and Tatiana F. Vieira and Clémence Richetta and Frédéric Subra and Samuel Lebourgeois and Charlotte Charpentier and Diane Descamps and Benoît Visseaux and Pierre Weigel and Alexandre Favereaux and Claire Beauvineau and Frédéric Buron and Marie-Paule Teulade-Fichou and Sylvain Routier and Sarah Gallois-Montbrun and Laurent Meertens and Olivier Delelis and Sérgio F. Sousa and Vincent Parissi},
url = {https://journals.asm.org/doi/abs/10.1128/aac.00083-22
hal-03826873v1 },
doi = {10.1128/aac.00083-22},
year = {2022},
date = {2022-07-05},
urldate = {2022-07-05},
journal = {Antimicrobial Agents and Chemotherapy},
volume = {0},
number = {0},
pages = {e00083-22},
abstract = {Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the infectious agent that has caused the current coronavirus disease (COVID) pandemic. Viral infection relies on the viral S (spike) protein/cellular receptor ACE2 interaction. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the infectious agent that has caused the current coronavirus disease (COVID) pandemic. Viral infection relies on the viral S (spike) protein/cellular receptor ACE2 interaction. Disrupting this interaction would lead to early blockage of viral replication. To identify chemical tools to further study these functional interfaces, 139,146 compounds from different chemical libraries were screened through an S/ACE2 in silico virtual molecular model. The best compounds were selected for further characterization using both cellular and biochemical approaches, reiterating SARS-CoV-2 entry and the S/ACE2 interaction. We report here two selected hits, bis-indolyl pyridine AB-00011778 and triphenylamine AB-00047476. Both of these compounds can block the infectivity of lentiviral vectors pseudotyped with the SARS-CoV-2 S protein as well as wild-type and circulating variant SARS-CoV-2 strains in various human cell lines, including pulmonary cells naturally susceptible to infection. AlphaLISA and biolayer interferometry confirmed a direct inhibitory effect of these drugs on the S/ACE2 association. A specific study of the AB-00011778 inhibitory properties showed that this drug inhibits viral replication with a 50% effective concentration (EC50) between 0.1 and 0.5 μM depending on the cell lines. Molecular docking calculations of the interaction parameters of the molecules within the S/ACE2 complex from both wild-type and circulating variants of the virus showed that the molecules may target multiple sites within the S/ACE2 interface. Our work indicates that AB-00011778 constitutes a good tool for modulating this interface and a strong lead compound for further therapeutic purposes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2 publications
Depienne, Sébastien; Alvarez-Dorta, Dimitri; Croyal, Mikael; Temgoua, Ranil C T; Charlier, Cathy; Deniaud, David; Mével, Mathieu; Boujtita, Mohammed; Gouin, Sébastien G
Luminol anchors improve the electrochemical-tyrosine-click labelling of proteins Article de journal
Dans: Chem Sci, vol. 12, no. 46, p. 15374–15381, 2021, ISSN: 2041-6520.
@article{pmid34976358,
title = {Luminol anchors improve the electrochemical-tyrosine-click labelling of proteins},
author = {Sébastien Depienne and Dimitri Alvarez-Dorta and Mikael Croyal and Ranil C T Temgoua and Cathy Charlier and David Deniaud and Mathieu Mével and Mohammed Boujtita and Sébastien G Gouin},
url = {hal-03429234v2 },
doi = {10.1039/d1sc04809k},
issn = {2041-6520},
year = {2021},
date = {2021-12-01},
urldate = {2021-12-01},
journal = {Chem Sci},
volume = {12},
number = {46},
pages = {15374--15381},
abstract = {New methods for chemo-selective modifications of peptides and native proteins are important in chemical biology and for the development of therapeutic conjugates. Less abundant and uncharged amino-acid residues are interesting targets to form less heterogeneous conjugates and preserve biological functions. Phenylurazole (PhUr), -methylphenylurazole (NMePhUr) and -methylluminol (NMeLum) derivatives were described as tyrosine (Y) anchors after chemical or enzymatic oxidations. Recently, we developed the first electrochemical Y-bioconjugation method coined eY-click to activate PhUr in biocompatible media. In this work, we assessed the limitations, benefits and relative efficiencies of eY-click conjugations performed with a set of PhUr, NMePhUr and NMeLum derivatives. Results evidenced a high efficiency of NMeLum that showed a complete Y-chemoselectivity on polypeptides and biologically relevant proteins after soft electrochemical activation. Side reactions on nucleophilic or heteroaromatic amino-acids such as lysine or tryptophan were never observed during mass spectrometry analysis. Myoglobine, bovine serum albumin, a plant mannosidase, glucose oxidase and the therapeutically relevant antibody trastuzumab were efficiently labelled with a fluorescent probe in a two-step approach combining eY-click and strain-promoted azide-alkyne cyclization (SPAAC). The proteins conserved their structural integrity as observed by circular dichroism and the trastuzumab conjugate showed a similar binding affinity for the natural HER2 ligand as shown by bio-layer interferometry. Compared to our previously described protocol with PhUr, eY-click with NMeLum species showed faster reaction kinetics, higher (complete) Y-chemoselectivity and reactivity, and offers the interesting possibility of the double tagging of solvent-exposed Y.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Lapaillerie, Delphine; Charlier, Cathy; Fernandes, Henrique S; Sousa, Sergio F; Lesbats, Paul; Weigel, Pierre; Favereaux, Alexandre; Guyonnet-Duperat, Véronique; Parissi, Vincent
In Silico, In Vitro and In Cellulo Models for Monitoring SARS-CoV-2 Spike/Human ACE2 Complex, Viral Entry and Cell Fusion Article de journal
Dans: Viruses, vol. 13, no. 3, p. 365, 2021, ISSN: 1999-4915.
@article{lapaillerie_silico_2021,
title = {In Silico, In Vitro and In Cellulo Models for Monitoring SARS-CoV-2 Spike/Human ACE2 Complex, Viral Entry and Cell Fusion},
author = {Delphine Lapaillerie and Cathy Charlier and Henrique S Fernandes and Sergio F Sousa and Paul Lesbats and Pierre Weigel and Alexandre Favereaux and Véronique Guyonnet-Duperat and Vincent Parissi},
url = {https://www.mdpi.com/1999-4915/13/3/365},
doi = {10.3390/v13030365},
issn = {1999-4915},
year = {2021},
date = {2021-01-01},
urldate = {2021-04-30},
journal = {Viruses},
volume = {13},
number = {3},
pages = {365},
abstract = {Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent responsible for the recent coronavirus disease 2019 (COVID-19) pandemic. Productive SARS-CoV-2 infection relies on viral entry into cells expressing angiotensin-converting enzyme 2 (ACE2). Indeed, viral entry into cells is mostly mediated by the early interaction between the viral spike protein S and its ACE2 receptor. The S/ACE2 complex is, thus, the first contact point between the incoming virus and its cellular target; consequently, it has been considered an attractive therapeutic target. To further characterize this interaction and the cellular processes engaged in the entry step of the virus, we set up various in silico, in vitro and in cellulo approaches that allowed us to specifically monitor the S/ACE2 association. We report here a computational model of the SARS-CoV-2 S/ACE2 complex, as well as its biochemical and biophysical monitoring using pulldown, AlphaLISA and biolayer interferometry (BLI) binding assays. This led us to determine the kinetic parameters of the S/ACE2 association and dissociation steps. In parallel to these in vitro approaches, we developed in cellulo transduction assays using SARS-CoV-2 pseudotyped lentiviral vectors and HEK293T-ACE2 cell lines generated in-house. This allowed us to recapitulate the early replication stage of the infection mediated by the S/ACE2 interaction and to detect cell fusion induced by the interaction. Finally, a cell imaging system was set up to directly monitor the S/ACE2 interaction in a cellular context and a flow cytometry assay was developed to quantify this association at the cell surface. Together, these different approaches are available for both basic and clinical research, aiming to characterize the entry step of the original SARS-CoV-2 strain and its variants as well as to investigate the possible chemical modulation of this interaction. All these models will help in identifying new antiviral agents and new chemical tools for dissecting the virus entry step.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2 publications
Ayadi, Nizar; Lafont, Florian; Charlier, Cathy; Benhelli-Mokrani, Houda; Sokolov, Pavel; Sukhanova, Alyona; Fleury, Fabrice; Nabiev, Igor
Comparative Advantages and Limitations of Quantum Dots in Protein Array Applications Chapitre d'ouvrage
Dans: Quantum Dots, vol. 2135, p. 259–273, Springer, New York, NY, Humana, 2020.
@inbook{cEQ3:ayadi_FLEURY:2020,
title = {Comparative Advantages and Limitations of Quantum Dots in Protein Array Applications},
author = {Nizar Ayadi and Florian Lafont and Cathy Charlier and Houda Benhelli-Mokrani and Pavel Sokolov and Alyona Sukhanova and Fabrice Fleury and Igor Nabiev},
year = {2020},
date = {2020-04-01},
booktitle = {Quantum Dots},
volume = {2135},
pages = {259--273},
publisher = {Springer},
address = {New York, NY},
edition = {Humana},
series = {Methods in Molecular Biology},
keywords = {},
pubstate = {published},
tppubtype = {inbook}
}
Dussouy, Christophe; Téletchéa, Stéphane; Lambert, Annie; Charlier, Cathy; Botez, Iuliana; Ceuninck, Frédéric De; Grandjean, Cyrille
Access to Galectin-3 Inhibitors from Chemoenzymatic Synthons Article de journal
Dans: The Journal of Organic Chemistry, vol. 85, no. 24, p. 16099-16114, 2020, (PMID: 33200927).
@article{doi:10.1021/acs.joc.0c01927b,
title = {Access to Galectin-3 Inhibitors from Chemoenzymatic Synthons},
author = {Christophe Dussouy and Stéphane Téletchéa and Annie Lambert and Cathy Charlier and Iuliana Botez and Frédéric De Ceuninck and Cyrille Grandjean},
url = {https://doi.org/10.1021/acs.joc.0c01927},
doi = {10.1021/acs.joc.0c01927},
year = {2020},
date = {2020-01-01},
journal = {The Journal of Organic Chemistry},
volume = {85},
number = {24},
pages = {16099-16114},
abstract = {Chemoenzymatic strategies are useful for providing both regio- and stereoselective access to bioactive oligosaccharides. We show herein that a glycosynthase mutant of a Thermus thermophilus α-glycosidase can react with unnatural glycosides such as 6-azido-6-deoxy-d-glucose/glucosamine to lead to β-d-galactopyranosyl-(1→3)-d-glucopyranoside or β-d-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-d-glucopyranoside derivatives bearing a unique azide function. Taking advantage of the orthogonality between the azide and the hydroxyl functional groups, the former was next selectively reacted to give rise to a library of galectin-3 inhibitors. Combining enzyme substrate promiscuity and bioorthogonality thus appears as a powerful strategy to rapidly access to sugar-based ligands},
note = {PMID: 33200927},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1 publication
Velic, Denis; Charlier, Cathy; Popova, Milena; Jaunet-Lahary, Titouan; Bouchouireb, Zakaria; Henry, Sébastien; Weigel, Pierre; Masson, Jean-Yves; Laurent, Adèle D; Nabiev, Igor; Fleury, Fabrice
Interactions of the Rad51 inhibitor DIDS with human and bovine serum albumins: Optical spectroscopy and isothermal calorimetry approaches Article de journal
Dans: Biochimie, vol. 167, p. 187–197, 2019, ISSN: 0300-9084.
@article{VELIC2019187,
title = {Interactions of the Rad51 inhibitor DIDS with human and bovine serum albumins: Optical spectroscopy and isothermal calorimetry approaches},
author = {Denis Velic and Cathy Charlier and Milena Popova and Titouan Jaunet-Lahary and Zakaria Bouchouireb and Sébastien Henry and Pierre Weigel and Jean-Yves Masson and Adèle D Laurent and Igor Nabiev and Fabrice Fleury},
url = {http://www.sciencedirect.com/science/article/pii/S0300908419302743},
doi = {https://doi.org/10.1016/j.biochi.2019.09.016},
issn = {0300-9084},
year = {2019},
date = {2019-01-01},
journal = {Biochimie},
volume = {167},
pages = {187--197},
abstract = {Rad51 is a key protein in DNA repair by homologous recombination and an important target for development of drugs in cancer therapy. 4′-diisothiocyanostilbene-2,2′-disulfonic acid (DIDS) has been used in clinic during the past 30 years as an inhibitor of anion transporters and channels. Recently DIDS has been demonstrated to affect Rad51-mediated homologous pairing and strand exchange, key processes in homologous recombination. Consequently, DIDS has been considered as a potential revertant of radio- and chemo-resistance of cancer cells, the major causes of therapy failure. Here, we have investigated the behavior of DIDS towards serum albumins. The effects of environmental factors, primarily, solvent polarity, on DIDS stability were evaluated, and the mechanisms of interaction of DIDS with human or bovine serum albumin were analyzed using isothermal calorimetry, circular dichroism and fluorescence spectroscopies. DIDS interaction with both serum albumins have been demonstrated, and the interaction characteristics have been determined. By comparing these characteristics for several DIDS derivatives, we have identified the DIDS moiety essential for the interaction. Furthermore, site competition data indicate that human albumin has two DIDS-binding sites: a high-affinity site in the IIIA subdomain and a low-affinity one in the IB subdomain. Molecular docking has revealed the key molecular moieties of DIDS responsible for its interactions in each site and shown that the IB site can bind two ligands. These findings show that binding of DIDS to serum albumin may change the balance between the free and bound DIDS forms, thereby affecting its bioavailability and efficacy against Rad51.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1 publication
Lafont, Florian; Ayadi, Nizar; Charlier, Cathy; Weigel, Pierre; Nabiev, Igor; Benhelli-Mokrani, Houda; Fleury, Fabrice
Assessment of DNA-PKcs kinase activity by quantum dot–based microarray Article de journal
Dans: Scientific Reports, vol. 8, no. 1, p. 1–12, 2018, ISSN: 20452322.
@article{Lafont2018,
title = {Assessment of DNA-PKcs kinase activity by quantum dot–based microarray},
author = {Florian Lafont and Nizar Ayadi and Cathy Charlier and Pierre Weigel and Igor Nabiev and Houda Benhelli-Mokrani and Fabrice Fleury},
doi = {10.1038/s41598-018-29256-2},
issn = {20452322},
year = {2018},
date = {2018-01-01},
journal = {Scientific Reports},
volume = {8},
number = {1},
pages = {1--12},
abstract = {Therapeutic efficacy against cancer is often based on a variety of DNA lesions, including DNA double-strand breaks (DSBs) which are repaired by homologous recombination and non-homologous end joining (NHEJ) pathways. In the past decade, the functions of the DNA repair proteins have been described as a potential mechanism of resistance in tumor cells. Therefore, the DNA repair proteins have become targets to improve the efficacy of anticancer therapy. Given the central role of DNA-PKcs in NHEJ, the therapeutic efficacy of targeting DNA-PKcs is frequently described as a strategy to prevent repair of treatment-induced DNA damage in cancer cells. The screening of a new inhibitor acting as a sensitizer requires the development of a high-throughput tool in order to identify and assess the most effective molecule. Here, we describe the elaboration of an antibody microarray dedicated to the NHEJ pathway that we used to evaluate the DNA-PKcs kinase activity in response to DNA damage. By combining a protein microarray with Quantum-Dot detection, we show that it is possible to follow the modification of phosphoproteomic cellular profiles induced by inhibitors during the response to DNA damage. Finally, we discuss the promising tool for screening kinase inhibitors and targeting DSB repair to improve cancer treatment.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1 publication
Bosseboeuf, Adrien; Feron, Delphine; Tallet, Anne; Rossi, Cédric; Charlier, Cathy; Garderet, Laurent; Caillot, Denis; Moreau, Philippe; Cardó-Vila, Marina; Pasqualini, Renata; Arap, Wadih; Nelson, Alfreda Destea; Wilson, Bridget S; Perreault, Hélène; Piver, Eric; Weigel, Pierre; Girodon, François; Harb, Jean; Bigot-Corbel, Edith; Hermouet, Sylvie
Monoclonal IgG in MGUS and multiple myeloma targets infectious pathogens Article de journal
Dans: JCI Insight, vol. 2, no. 19, p. 1–18, 2017, ISSN: 0021-9738.
@article{Bosseboeuf2017,
title = {Monoclonal IgG in MGUS and multiple myeloma targets infectious pathogens},
author = {Adrien Bosseboeuf and Delphine Feron and Anne Tallet and Cédric Rossi and Cathy Charlier and Laurent Garderet and Denis Caillot and Philippe Moreau and Marina Cardó-Vila and Renata Pasqualini and Wadih Arap and Alfreda Destea Nelson and Bridget S Wilson and Hélène Perreault and Eric Piver and Pierre Weigel and François Girodon and Jean Harb and Edith Bigot-Corbel and Sylvie Hermouet},
doi = {10.1172/jci.insight.95367},
issn = {0021-9738},
year = {2017},
date = {2017-01-01},
journal = {JCI Insight},
volume = {2},
number = {19},
pages = {1--18},
abstract = {Subsets of mature B cell neoplasms are linked to infection with intracellular pathogens such as Epstein-Barr virus (EBV), hepatitis C virus (HCV), or Helicobacter pylori. However, the association between infection and the immunoglobulin-secreting (Ig-secreting) B proliferative disorders remains largely unresolved. We investigated whether the monoclonal IgG (mc IgG) produced by patients diagnosed with monoclonal gammopathy of undetermined significance (MGUS) or multiple myeloma (MM) targets infectious pathogens. Antigen specificity of purified mc IgG from a large patient cohort (n = 244) was determined using a multiplex infectious-antigen array (MIAA), which screens for reactivity to purified antigens or lysates from 9 pathogens. Purified mc IgG from 23.4% of patients (57 of 244) specifically recognized 1 pathogen in the MIAA. EBV was the most frequent target (15.6%), with 36 of 38 mc IgGs recognizing EBV nuclear antigen-1 (EBNA-1). MM patients with EBNA-1-specific mc IgG (14.0%) showed substantially greater bone marrow plasma cell infiltration and higher β2-microglobulin and inflammation/infection-linked cytokine levels compared with other smoldering myeloma/MM patients. Five other pathogens were the targets of mc IgG: herpes virus simplex-1 (2.9%), varicella zoster virus (1.6%), cytomegalovirus (0.8%), hepatitis C virus (1.2%), and H. pylori (1.2%). We conclude that a dysregulated immune response to infection may underlie disease onset and/or progression of MGUS and MM for subsets of patients.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1 publication
Forato, Florian; Liu, Hao; Benoit, Roland; Fayon, Franck; Charlier, Cathy; Fateh, Amina; Defontaine, Alain; Tellier, Charles; Talham, Daniel R; Queffélec, Clémence; Bujoli, Bruno
Comparison of Zirconium Phosphonate-Modified Surfaces for Immobilizing Phosphopeptides and Phosphate-Tagged Proteins Article de journal
Dans: Langmuir, vol. 32, no. 22, p. 5480–5490, 2016, ISSN: 15205827.
@article{Forato2016,
title = {Comparison of Zirconium Phosphonate-Modified Surfaces for Immobilizing Phosphopeptides and Phosphate-Tagged Proteins},
author = {Florian Forato and Hao Liu and Roland Benoit and Franck Fayon and Cathy Charlier and Amina Fateh and Alain Defontaine and Charles Tellier and Daniel R Talham and Clémence Queffélec and Bruno Bujoli},
doi = {10.1021/acs.langmuir.6b01020},
issn = {15205827},
year = {2016},
date = {2016-01-01},
journal = {Langmuir},
volume = {32},
number = {22},
pages = {5480--5490},
abstract = {Different routes for preparing zirconium phosphonate-modified surfaces for immobilizing biomolecular probes are compared. Two chemical-modification approaches were explored to form self-assembled monolayers on commercially available primary amine-functionalized slides, and the resulting surfaces were compared to well-characterized zirconium phosphonate monolayer-modified supports prepared using Langmuir-Blodgett methods. When using POCl3 as the amine phosphorylating agent followed by treatment with zirconyl chloride, the result was not a zirconium-phosphonate monolayer, as commonly assumed in the literature, but rather the process gives adsorbed zirconium oxide/hydroxide species and to a lower extent adsorbed zirconium phosphate and/or phosphonate. Reactions giving rise to these products were modeled in homogeneous-phase studies. Nevertheless, each of the three modified surfaces effectively immobilized phosphopeptides and phosphopeptide tags fused to an affinity protein. Unexpectedly, the zirconium oxide/hydroxide modified surface, formed by treating the amine-coated slides with POCl3/Zr4+, afforded better immobilization of the peptides and proteins and efficient capture of their targets.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}